RESEARCH INTERESTS
B. AIDS Cure
C. New restriction factors to HIV replication
D. AIRE
A. Transcription and co-transcriptional processing in eukaryotes
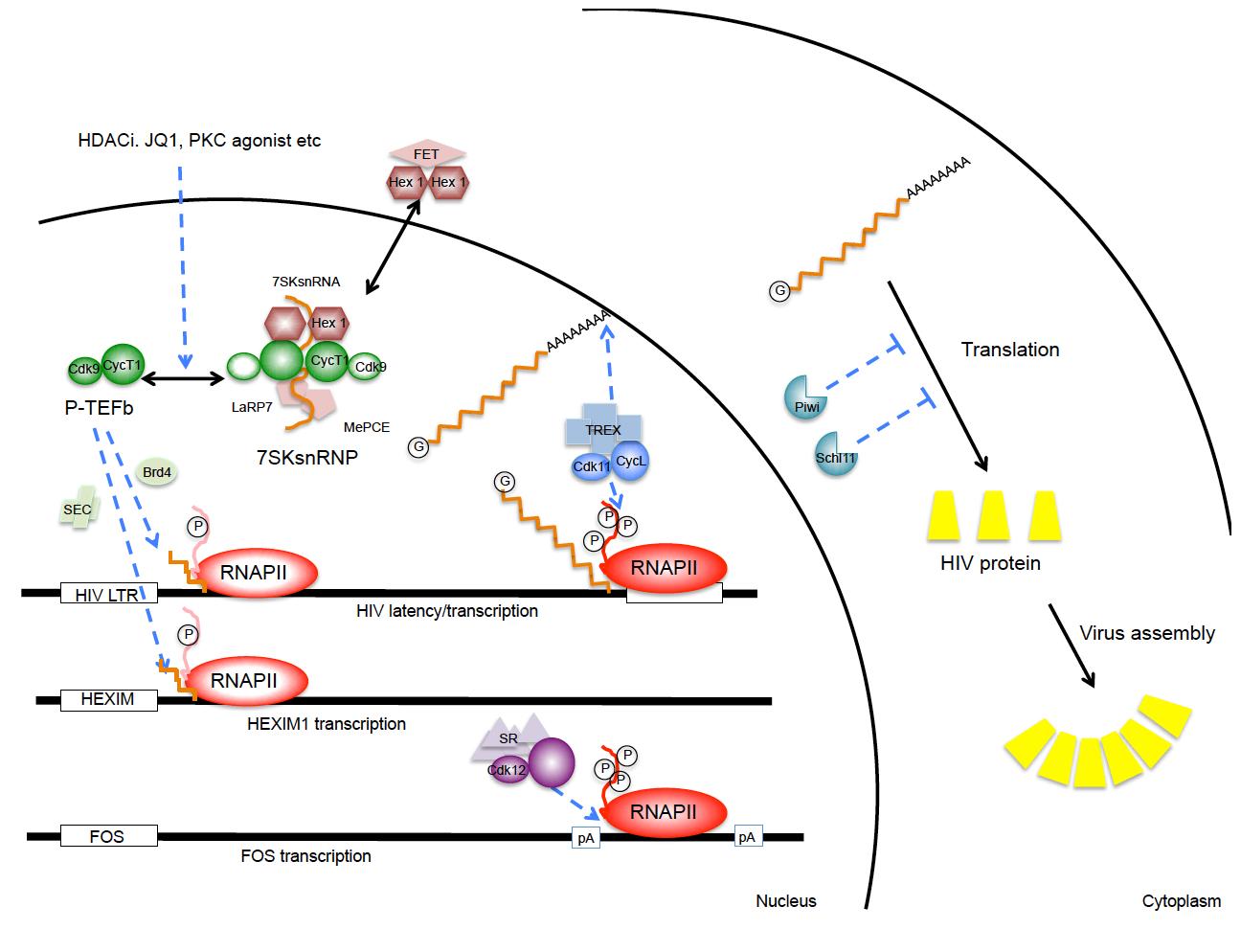
At least three separate cyclin-dependent kinases help to regulate eukaryotic gene expression at levels of elongation, splicing, 3’ end formation and polyadenylation (Fig.1). They are CycT1/T2:CDK9, CycL1/L2:CDK11 and CyK:CDK12/13. CycT1/CKD9 is also known as the positive transcription elongation factor b (P-TEFb). It is required to release stalled RNA polymerases II (RNAPII) at promoters for productive elongation. P-TEFb phosphorylates negative elongation factors (DSIF and NELF) and serines at position 2 (S2P) in the C-terminal domain (CTD) of RNAPII, converting DSIF into an elongation factor and allowing RNAPII to assemble splicing and cleavage/polyadenylation factors. P-TEFb can be recruited to promoters via activators such as NF-kB, cMyc, CIITA, steroid hormone receptors, etc, BRD4, the super elongation complex (SEC), or in the case of HIV, by its transactivator Tat. Levels of P-TEFb are low in resting cells, but increase following cellular activation and proliferation signals. In cycling cells, P-TEFb partitions between an active (free P-TEFb) and an inactive (7SK snRNP) form. This P-TEFb equilibrium determines the state of activation and proliferation of the cell. Since 7SK snRNA is required to assemble the inactive form, its integrity is paramount and depends on MePCE and LaRP7, which bind and stabilize its 5’ and 3’ ends, respectively. Many tumors contain mutations in LaRP7 and thus do not maintain a proper P-TEFb equilibrium. In our studies on P-TEFb, we discovered that most differentiation agents, HDAC and BET bromodomain inhibitors act via a transient release of P-TEFb from the 7SK snRNP, which is followed rapidly by the increased synthesis of HEXIM1 that binds to 7SK snRNA and inactivates P-TEFb. Thus, the reassembly of the 7SK snRNP is rapid and most P-TEFb is inactivated, thus resulting in growth arrest and/or differentiation of the cell. For purposes of drug discovery, we have now developed a rapid test to follow changes in the P-TEFb equilibrium that correlates directly with changes in the physiology of the cell. Structural studies of the 7SK snRNP are ongoing.
CycL/CDK11 associates with the TREX/THOC complex and is thus recruited to RNAPII. It is another CTD kinase, whose effects are primarily on polyadenylation and stability, export and translation of mRNA species. In our studies, elevated levels of CycL/CDK11 increased dramatically the expression of HIV. Again, levels of this complex are low in resting cells, which contribute to silencing of host cell and HIV gene expression. Ongoing studies are dissecting structural and functional aspects of this complex regulatory mechanism.
CycK/CDK12 associates with the splicing machinery and ensures proper 3’ end formation of long genes, such as those that regulate responses to dsDNA damage, BRCA1/2, ATM, ATR, etc. Again, this CTD kinase ensures that transcripts are cleaved and polyadenylated efficiently. CycK/CDK12 also ensures that acute response genes prefer proximal to distal polyadenylation sites. Ongoing studies are similar to those with CycL/CDK11 and are designed to reveal differences and similarities between the two systems. They might reveal some secrets of the CTD code and how some processing/polyadenylation factors are preferentially recruited by different genes and upon different stimuli.
B. AIDS cure
P-TEFb and CycL/CDK11 are both essential for HIV transcription and replication. Their low levels in resting cells help to maintain proviral latency in the host, i.e. ensure that viral transcripts are not expressed. A major focus of the laboratory is to increase levels of these CTD kinases and NF-kB to reactivate HIV replication in this reservoir so that optimal anti-retroviral therapy (ART), the immune system and direct cell killing by the virus eliminate HIV from the infected host. To this end, we already defined drugs that could be used in clinical trials. We are also very interested in how transcriptional interference drives HIV into latency and what approaches also reactivate the virus in these cells. In short, we established paradigms and validated approaches to shock and kill HIV-infected cells in optimally treated individuals. We also established visual screening approaches for finding new drugs that could be used for the cure of AIDS in the future.
|
|
|
Fig.1. Involvement of CTD kinases and restriction factors in HIV replication. Depicted are P-TEFb, CycL:CDK11 and CycK:CDK12 complexes. Release of P-TEFb from the 7SK snRNP occurs following many stimuli that stress cells. PKC agonists, HDAC and BET bromodomain inhibitors can achieve the same release. CycL:CDK11 and CycK:CDK12 affect 3’ end formation and polyadenylation of HIV and other transcripts. Levels of these CTD kinases are low in resting cells, which contributes to proviral latency in the infected host. In the cytoplasm, Schlafen 11 and Piwil proteins can inhibit translation, i.e. protein expression of HIV and other retroviruses/retrotransposons. |
C. New cellular restriction factors for HIV, other viruses and retrotransposons.
We are studying cellular proteins that interfere with HIV translation, such as Schlafen 11, Ago2/RISC and Piwil proteins (Fig.1). To these ends, the suboptimal codon usage of HIV and other retroviruses, some dictated by the extensive secondary structures of their genomic RNAs, some resulting from extensive editing by APOBEC proteins, render these pathogens very susceptible to levels of tRNAs in cells. Thus, some of these small ncRNA-binding proteins that interfere with retroviral translation are tRNAs that are being depleted rather than miRNAs or siRNAs. These translational problems might also play important roles in restricting retrotransposition during germ line development and spermatogenesis as well as maternal-fetal transmission of HIV.
D. AIRE
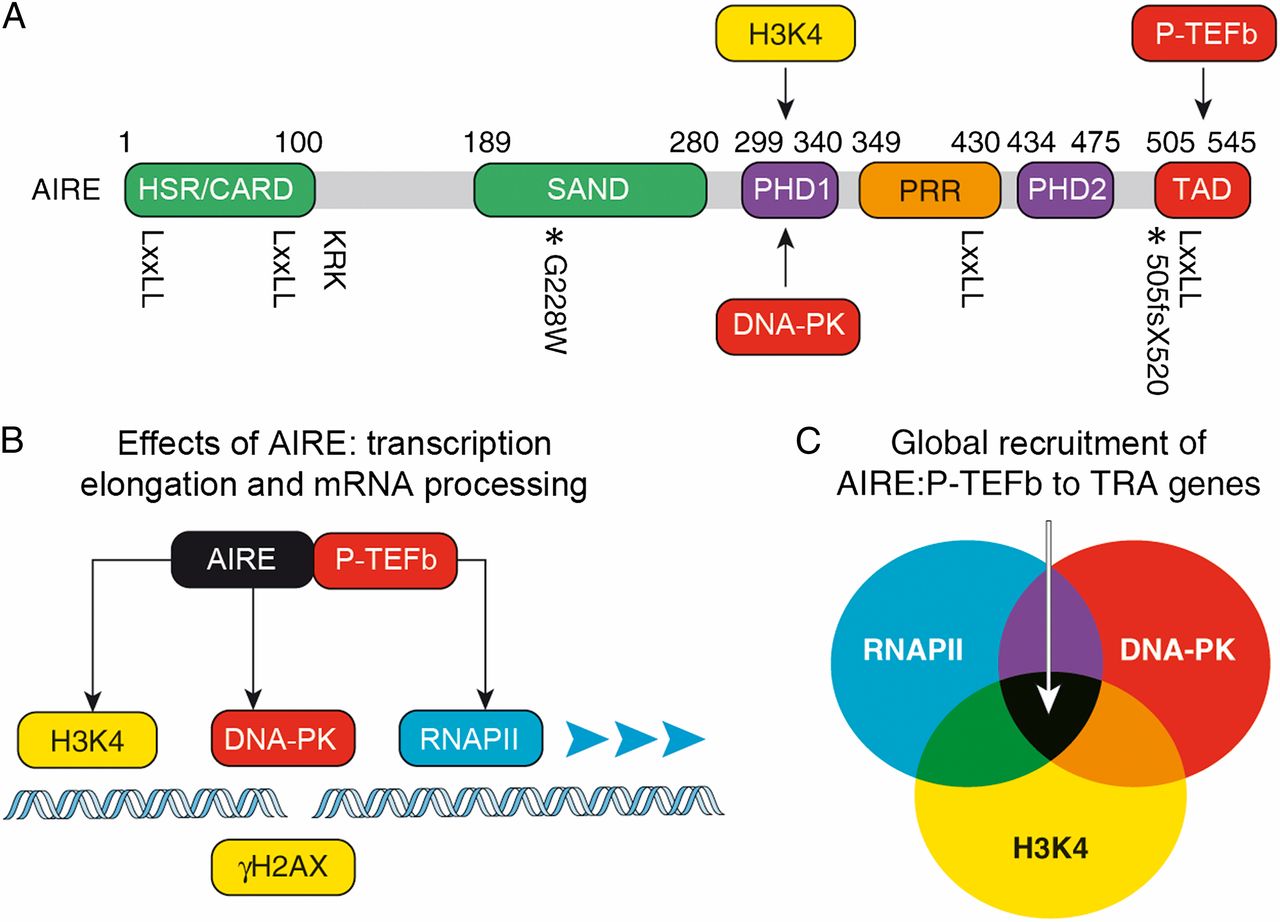
The autoimmune regulator AIRE is a transcription factor that dictates central tolerance in the thymus and helps to maintain tolerance in the periphery. It activates otherwise tissue restricted antigen (TRA) genes in specialized epithelial cells. Their peptides are presented on major histocompatibility complex class II (MHCII) determinants to eliminate self-reacting developing T cells in the thymus. They also contribute to anergy of mature T cells via extrathymic AIRE-expressing cells in lymph nodes and spleen. We are interested in AIRE because it activates otherwise silent genes in cells. It is recruited there via unmodified histone H3 (H3K4), DNA-PK and RNAPII (Fig.2). Since chromatin is not modified at these sites and along genes to be transcribed, we are investigating if epigenetic changes occur prior to or after transcription. Thus, the central question is whether chromatin modifications modify transcription or whether transcription and RNAPII modify chromatin. We favor the notion that most epigenetic changes on genes result from active transcription and not vice versa.
|
|
|
Fig.2. AIRE protein domains, key interacting partners, and mechanism of TRA gene expression. (A) AIRE contains several domains that are related to those in other transcription factors. From the N terminus they are: HSR (green), which is important for the oligomerization of AIRE and may function as a caspase recruitment domain; SAND (green); PHD1 and PHD2 (violet); proline-rich region (PRR; orange); and the TAD (red). Protein residues corresponding to human AIRE are labeled above, and the position of the four LXXLL motifs is marked below the diagram. The location of the nuclear localization signal is also indicated (KRK). Relevant mutations are marked with an asterisk below the diagram with accompanying labels. Arrows depict interactions between DNA-PK, unmodified histone H3 (H3K4), P-TEFb, and AIRE. (B) Schematic diagram depicting the molecular mechanism of AIRE-regulated TRA gene expression. Combinatorial interactions between AIRE, unmodified H3K4 (yellow), DNA-PK (red), and RNAPII (blue) recruit AIRE to a TRA promoter. AIRE brings P- TEFb (red) to phosphorylate the RNAPII CTD, and this results in transcription elongation, mRNA processing, and TRA gene expression. Histone H2AX with phosphorylated S139 (gH2AX), which marks DNA double-strand breaks, is depicted in yellow. (C) The Venn diagram marks sets of genes that contain unmodified H3K4, engaged RNAPII, and DNA-PK at their promoters. AIRE is targeted to and can regulate expression of genes at the intersection of these sets. |